
Generic advice about setting up and using conda environments
Source:vignettes/setup_and_tips.Rmd
setup_and_tips.RmdIntroduction
SeuratIntegrate’s main purpose is to extend the range of
scRNA-seq integration tools available in R and compatible with
Seurat. Many of them being solely available in Python, we
developed wrappers leveraging reticulate package’s
capabilities. reticulate enables to directly call Python
from R, give that conda environments have been set up beforehand.
The purpose of this vignette is to illustrate and ease the installation and the use of those environments.
Prior to starting, make sure you have:
- conda installed on your machine
- conda binary in your
PATH(or that you know its location)
Set-up
We need multiple conda environments. One for bbknn, one
for Scanorama, one for
scVI/scANVI and one for trVAE. If
you don’t plan on using any or some of these methods, whether you decide
to set up their respective conda environments is up to you.
One the contrary, if you already have some conda environments with
appropriate libraries on your machine, you can tell
SeuratIntegrate to use them. Let’s see how.
Have look at your CondaEnvManager:
getCache()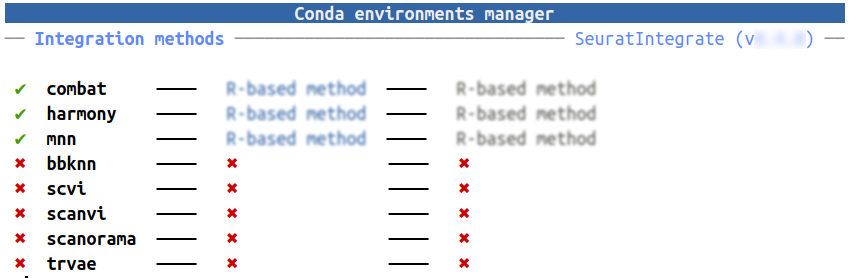
It’s a handy way to have a glance at all the implemented methods and
the status of their conda environments. ComBat,
Harmony and MNN are R-based methods and don’t
need any conda environment. The rest of them however are Python-based
and will function through reticulate, hence require conda
environments.
If you don’t have any conda environments for them, look at the next sub-section. Conversely, if you want to add an existing conda environment, directly go to the following one.
Option 1: Create new conda environments with
SeuratIntegrate
Note that the commands below have only been tested on Linux distributions
Try the following commands (be aware that execution might take time):
UpdateEnvCache('bbknn')
UpdateEnvCache('scvi')
UpdateEnvCache('scanorama')
UpdateEnvCache('trvae')Note that:
- if conda is not in you PATH set
conda.bin = /path/to/conda -
scVIandscANVIshare the same environment. Hence, it is not necessary to run bothUpdateEnvCache('scvi')andUpdateEnvCache('scanvi')
Have look at your CondaEnvManager:
getCache()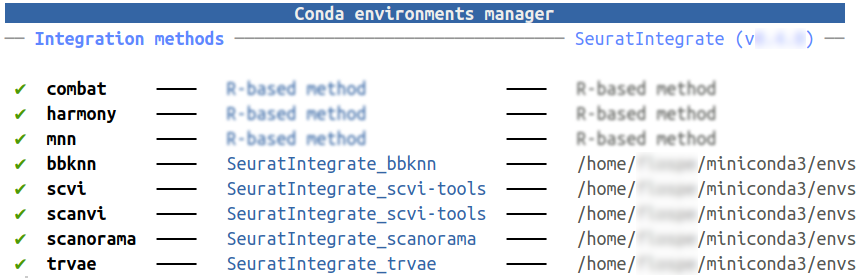
Option 2: Use existing conda environments
If you already have one (several) existing conda environment(s) for
one (some) of the methods, you can tell SeuratIntegrate to
use it (them). Similarly, if you run into problems with
UpdateEnvCache() commands above, the alternative is to set
up conda environments yourself and provide them to
SeuratIntegrate. Whatever the case, let’s proceed.
You’ll use UpdateEnvCache(). You can specify the name of
the conda environment or the path to it. By default,
UpdateEnvCache() will try to decide whether the provided
value for conda.env is a path or a name based on simple
tests. To avoid any misinterpretation, you can use
conda.env.is.path = TRUE or FALSE when your
input is the path or the name of the environment, respectively.
But beware not to make mistakes !!!
See examples below. You should adapt the arguments to your situation:
# environment for bbknn
UpdateEnvCache('bbknn', conda.env = 'bbknn_env',
conda.env.is.path = FALSE) # default 'auto' would work
# environment for bbknn in ./bbknn_env/
UpdateEnvCache('bbknn', conda.env = 'bbknn_env',
conda.env.is.path = TRUE)
# environment for bbknn, conda binary not in PATH
UpdateEnvCache('bbknn', conda.env = 'bbknn_env', conda.bin = 'cutom/location/conda')
# path for scvi-tools
UpdateEnvCache('scvi', conda.env = '~/miniconda3/envs/scvi-tools_env',
conda.env.is.path = TRUE) # default 'auto' would workNote that:
- if conda is not in you PATH set
conda.bin = /path/to/conda - the
conda.binmust correspond to the conda managing theconda.env -
scVIandscANVIshare the same environment. Hence, it is not necessary to run bothUpdateEnvCache('scvi')andUpdateEnvCache('scanvi')
Now you can use the Python-based methods !
Update or reset conda environments
If you want to update a conda environment, use
# change 'method' by the name of the method
UpdateEnvCache(method = 'method', overwrite.env = TRUE, ...)To unset an environment, use
# change 'method' by the name of the method
resetCache(method = 'method')Troubleshouting with conda
Common issues with setting up environments
It can happen that a conda environment cannot be installed on a specific machine or os. In this case, there is hardly anything better to do than browse the internet in hope that someone else has experienced a similar problem and that the solution is public. Otherwise, try to modify the set of packages to install, be more or less stringent with package versions to install, etc. You can also create a conda environment with Python and pip, and then to try to install packages with pip rather than conda.
Once the problem is solved (if it can be), you can save the new
environment to the CondaEnvManager with
# change 'method' by the name of the method
# change'difficult_conda_env' by the name of the working conda environment
UpdateEnvCache(method = 'method', conda.env = 'difficult_conda_env')Possible issues with activating environments
It can happen that a conda environment does not work (or stops working) on a specific machine. Below are some potential causes of conflicts between Python libraries, conda itself or other components that could lead to malfunctions (non exhaustive list):
- with rmarkdown/knitr
How to check
Check if the same command works outside of an rmarkdown (e.g. in a R script), once you have closed any Rmardown file and closed the R session (and restrated RStudio). This is something to consider notably when you encounter an error like:
ImportError: /opt/conda/envs/[env_name]/lib/python3.10/site-packages/PIL/../../../libtiff.so.6: undefined symbol: jpeg12_write_raw_data, version LIBJPEG_8.0How to fix
Close any Rmardown file in the source panel, quit the R session (close RStudio) and work from a R script.- with RStudio
How to check
Check if the same command works outside of RStudio. For instance, if there is an error during an integration because scanpy cannot be imported, try:
# from a terminal
RHow to fix
Lunch the integration outside of RStudio (from a terminal for example).- with reticulate
How to fix
First, try to update reticulate. If it doesn’t work any better, check if someone has encountered the same issue (browse the web or the issues on the reticulate github repository. If nothing works, either post an issue on the reticulate github repos or retry to import after you have installed different Python package versions.
If this is a problem withmkl, install the conda
environment again with the arguments nomkl (and
-c conda-forge if not already).
- between Python packages
How to check
This is more tricky. But some packages are know to be incompatible.
For instance, jax and jaxlib work better when their versions are
identical. In my experience, the scvi-tools environment can
be set up with two discrepant versions of jax and jaxlib. To check,
try:
conda list -n [conda_env_name] jaxHow to fix
If this is a problem with jax and jaxlib, try to force install them with identical versions. Otherwise, search your error on the web.- between your local os and the list of packages to be installed by SeuratIntegrate
How to check
This is more an installation problem usually. Check the previous section.How to fix
This is more an installation problem usually. Check the previous section.Subtleties of using Future
If the following bullet points seem obscure, further explanations are given in the sections below. In brief,
- disable future for R-based methods (
DoIntegrate([...], use.future = FALSE))- never use
CCAandRPCAintegration methods with multisession (previous advice prevents this, especially for Windows users)- multicore futures are faster and less memory-intensive than multisession, but unstable on RStudio and unavailable on Windows
- to force
DoIntegrate()to use a multicore framework (at your own risk), setoptions(parallelly.fork.enable = FALSE). Unavailable on Windows
Why SeuratIntegrate uses Future ?
A R session can only initialise one Python environment at a time via
reticulate. This known
limitation
of reticualte is overcome by launching a “background
session” using Future. The environment is initialised
there instead of the main user’s R session. This feature is embedded in
DoIntegrate().
Futures are therefore useless for R-based methods and should be
disabled with DoIntegrate([...], use.future = FALSE).
Worse, it is discouraged with
CCAIntegration and RPCAIntegration
(explanations are in the final part of the vignette)
Inconveniences
In the vast majority of cases, the impact of the “futures” is insignificant. The most obvious are the few needed seconds to launch the future session and export globals, in addition to the reordering of stdout and message output, resulting in messy and less informative print statements intended for the user.
Tweaks
The package implements sequential, multicore, multisession, and
cluster futures. SeuratIntegrate only uses the multicore
and multisession ones. DoIntegrate() automatically picks
the preferred one based on the output of
parallelly::supportsMulticore()## [1] FALSEHere multicore is not supported, thus DoIntegrate()
would start a multisession. Further explanations regarding settings
giving priority to multicore are available in the function’s help (the
important part is in the disclosure widget
?supportsMulticore)
help('supportsMulticore', package = 'parallelly')?supportsMulticore
Support for process forking
While R supports forked processing on Unix-like operating system such as Linux and macOS, it does not on the Microsoft Windows operating system.
For some R environments it is considered unstable to perform parallel processing based on forking. This is for example the case when using RStudio, cf. RStudio Inc. recommends against using forked processing when running R from within the RStudio software. This function detects when running in such an environment and returns
FALSE, despite the underlying operating system supports forked processing. A warning will also be produced informing the user about this the first time time this function is called in an R session. This warning can be disabled by setting R optionparallelly.supportsMulticore.unstable, or environment variable R_PARALLELLY_SUPPORTSMULTICORE_UNSTABLE to"quiet".Enable or disable forked processing
It is possible to disable forked processing for futures by setting R option
parallelly.fork.enabletoFALSE. Alternatively, one can set environment variable R_PARALLELLY_FORK_ENABLE tofalse. Analogously, it is possible to override disabled forking by setting one of these toTRUE.
In a nutshell, multicore is
- unavailable on Windows
- discouraged (considered unstable) in some R environments, notably RStudio
- always used on Unix-like operating systems when
options(parallelly.fork.enable = TRUE)
The main reason for using multicore is that FORKing is considered to be faster and result in a lower memory overhead than PSOCK (i.e. multisession) (see there for technical details)
Furthermore, DoIntegrate() uses not only future, but
also NSE (non-standard evaluation), enabling to specify arguments within
each integration function call. Briefly, each call inside
DoIntegrate() (such as
DoIntegrate(bbknnIntegration())) is not directly executed
but captured to be executed later on (so that the proper value
for the object parameter can be passed on to
bbknnIntegration for instance). Further details are
available in the book Advanced
R by Hadley Wickham.
The important part is that, unlike for multicore, on a multisession
future, DoIntegrate() evaluates each argument before
launching the integration on the background R session. Thus, a Seurat
Assay object is passed as a structure (output of
str(object)). This takes time and makes the call extremely
long.
It has a unexpected side-effect. It slows down
CCAIntegration and RPCAIntegration a lot when
they are input with Seurat objects normalised with
SCTransform(). Indeed, they both call
FindIntegrationAnchors() -> merge() ->
merge.SCTAssay(). The latter performs a grep
on the previous calls (output of sys.calls()). In a
multisession future, big objects such as the Seurat Assay object are
passed as a structure and the grep can be
unnecessarily time-consuming. To avoid this, one can either specify
use.future = FALSE for the R-based method (this is always
preferable) or at least ban the use of CCA and
RPCA integrations with a multisession future (note that
Windows users can only pick the first option).