SeuratIntegrate streamlines single-cell transcriptomics (scRNA-seq) data integration and batch effect correction. This R package effortlessly extends the Seurat workflow with 8 popular integration methods across R and Python, complemented by 11 robust scoring metrics to estimate their performance.
Integrations
- ComBat
- Harmony
- MNN
- BBKNN
- scVI
- scANVI
- Scanorama
- trVAE
Scoring metrics
- ARI
- ASW
- Batch ASW
- Cell cycle conservation
- Graph connectivity
- PCA-density
- PCA-regression
- kBET
- cell-type (c)LISI
- batch (i)LISI
- NMI
Installation
Install SeuratIntegrate from github:
install.packages(c("remotes", "BiocManager")) # if not installed
remotes::install_github("cbib/Seurat-Integrate", repos = BiocManager::repositories()) To benefit from SeuratIntegrate’s full capabilities, we recommend installing the following packages:
# fast distance computation
install.packages('distances')
# required to test for k-nearest neighbour batch effects
remotes::install_github('theislab/kBET')
# faster Local Inverse Simpson’s Index computation
remotes::install_github('immunogenomics/lisi')Conda environments for Python methods
To simplify the creation and management of conda environments, we suggest using UpdateEnvCache():
# create environments:
UpdateEnvCache("bbknn")
UpdateEnvCache("scvi")
UpdateEnvCache("scanorama")
UpdateEnvCache("trvae")Those environments will be saved and automatically used by the Python integration methods provided by SeuratIntegrate.
Alternatively, the cache can be updated with a pre-existing environment. This can be useful if you have to set up a conda environment yourself because a command above failed or a conda environment turned out to be non-functional.
# save "my_bbknn_env" (for bbknn) to cache
UpdateEnvCache("bbknn", conda.env = "my_bbknn_env")The cache remains persistent across sessions and its state can be displayed with:
getCache()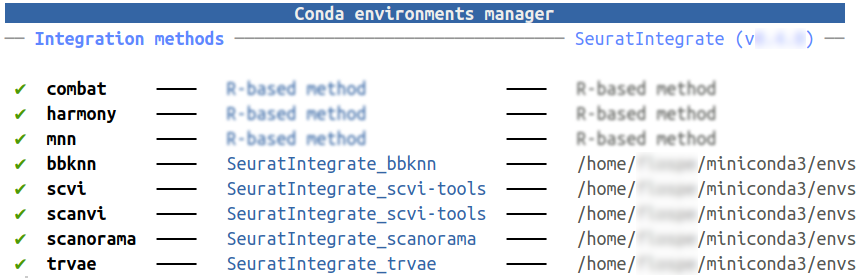
CondaEnvManager
Further details are provided in the vignette("setup_and_tips").
Data integration
To run integration algorithms, we have developed a function called DoIntegrate() that enables:
- performing multiple integration methods in a single call
- control over the data (raw, normalised or scaled) and the features to use as input
- flexible customisation of parameters for each integration method
seu <- DoIntegrate(seu,
# ... integrations
CombatIntegration(layers = "data"),
HarmonyIntegration(orig = "pca", dims = 1:30),
ScanoramaIntegration(ncores = 4L, layers = "data"),
scVIIntegration(layers = "counts", features = Features(seu)),
# ...
use.hvg = TRUE, # `VariableFeatures()`
use.future = c(FALSE, FALSE, TRUE, TRUE)
)Here, all integration methods will use the variable features as input, with the exception of scVIIntegration() which is set to use all features. CombatIntegration() will correct the normalised counts, while scVIIntegration() will train on raw counts.
use.future must be TRUE for Python methods, and FALSE for R methods.
Integration methods produce one or several outputs. Those can be of multiple types - either a new assay with corrected counts, a new dimension reduction with corrected cell embeddings, or a new graph with corrected edges. The type of output is important to consider, because it will require different post-processing steps until the result of the integration can be visualised on a UMAP:
- Corrected counts:
ScaleData()->RunPCA()->RunUMAP() - Dimension reduction:
RunUMAP() - KNN graph:
RunUMAP(umap.method = "umap-learn")
Performance assessment
SeuratIntegrate incorporates 11 scoring metrics: 6 quantify the degree of batch mixing (batch correction), while 5 assess the preservation of biological differences (bio-conservation) based on ground truth cell type labels.
Each score can be obtained using a function of the form Score[score_name](), or directly saved in the Seurat object using the AddScore[score_name]() counterpart:
# save the score in a variable
rpca_score <- ScoreRegressPC(seu, reduction = "pca")
# or save the score in the Seurat object
seu <- AddScoreRegressPC(seu, integration = "unintegrated", reduction = "pca")The AddScore functions have an advantage over the Score functions. They allow to then scale the scores between zero and one and to standardize their direction (the closer to one, always the better), facilitating their readability. Scores can eventually be plotted to readily compare the different integrations:
# scale
seu <- ScaleScores(seu)
# plot
PlotScores(seu)